Protein structures
Proteins with properties well-suited for their intended use are the products of molecular evolution processes that we study, whether they occur in Nature or by design in the laboratory. To further our understanding of the evolution processes themselves or the proteins' performance we have determined the structure of several such proteins, as briefly outlined below.
| PDB | PyMol view | Brief summary |
|---|---|---|
| 1AQK | 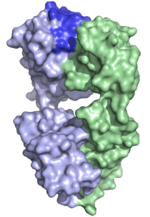 | This is a structure of a Fab fragment of a human monoclonal antibody specific for tetanus toxoid that is produced by a human × mouse heterohybridoma cell line. The binding site (on top) features a large aliphatic ridge dominating the center of the active site. The protein shows exceptional crystallization properties and has been proposed to be provide guidance for development of novel antibody constructs facilitating antibody fragment crystallization. (Abstract of the article) |
| 4BUH | 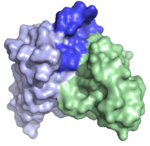 | This is a structure of an antibody fragment selected from a combinatorial antibody library that incorporates heavy chain variable domains of allergic subjects' IgE repertoire. It is specific for the birch pollen allergen Bet v 1. In contrast to previously crystallized IgEs, it features a very prominent protruding CDRH3 (dark blue) that extends well above the plane of the rest of the likely binding site. It thus represents the first example of such a human allergen-specific antibody. (Abstract of the article) |
| 3JXS | 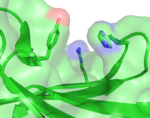 | This protein (XG-34) is a carbohydrate-bindning module evolved for specificity to a branched carbohydrate, non-fucosylated xyloglucan. The protein, the first of an evolved derivative of CBM4-2, retains the overall fold of CBM4-2 from which is was derived despite its substantially modified specificity. It features a cleft into which the saccharide likely fits, lined in particular by a histidine and tyrosine side chain. (Abstract of the article) In contrast to other modules derived from CBM4-2, it features an arginine side chain that extends into the cleft. In other modules this side chain instead make polar interactions with an adjacent side chain (E112) positioning it along the bottom of the cleft. This interaction is not possible in XG-34 as it has undergone a E112D mutation. Reversion of this mutation increases the affinity of the module for its ligand by approximately 2 orders of magnitude. (Abstract of the article) |
| 2Y6J | 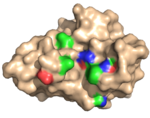 | This protein (X-2), also evolved from CBM4-2, is specific for xylan as it does not bind for instance β-glucan, xyloglucans or cellulose. It also features a cleft, in this case lined by a phenylalanine and a leucine residue in place of the tryptophane and phenylalaine residues found in the wild type protein sequence (it carries an additional six modifications, most of which are found in the bindning site (as illustrated in the accompanying figure) among its 165 residues in comparison to the wild type module). (Abstract of the article) |
| 2Y6L | 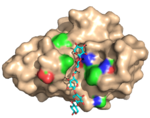 | X-2 bind oligoxylose (in this case the pentamer) in its cleft. The binding site fits all five units of this ligand. Multiple hydrophobic stacking interactions and polar interactions are lmade between the ligand and the protein. The structural adaptations that are required to accommodate the ligand are very limited indeed and the binding may be viewed upon as a lock-and-key binding mode. (Abstract of the article) |
| 2Y6K | 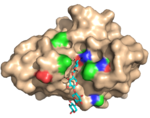 | X-2 binds the ligand tetramer (xylotetraose) in a manner very similar to the bindning of the pentamer allowing the ligand to occupy four of the five ligand interacting sub-sites. (Abstract of the article) |
| 2Y6H | 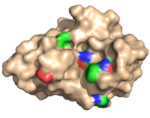 | X-2 L110F is a mutated variant of X-2 that has reversed the substitution of one of the residues (L110) that line the ligand binding cleft to the residue (phenylalanine) that is found in wild-type CBM4-2. X-2 L110F, in contrast to X-2, is, just as CBM4-2, highly ligand cross-reactive as a consequence of this substitution. The substitution, however, does not result in any major reorganization of the bindning site of the unligated module. (Abstract of the article) |
| 2Y64 | 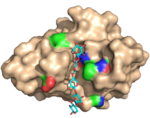 | X-2 L110F bind xylose oligosaccharides much in the same was as X-2 does through polar interactions and hydrophobic stacking. Very little structural adaptation is required to successfully bind this ligand. (Abstract of the article) |
| 2Y6G | 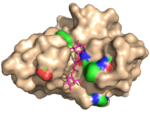 | X-2 L110F also bind oligomers of glucose, such as those found in cellulose. Substantial structural rearrangements (e.g. of aspartate 29 and arginine 142) of the CBM are, however, required to fit the ligand into the binding site for instance to accommodate some of the C6 hydroxymethyl groups of the ligand and to create new polar interactions. In addition, the ligand takes a partially different path through the binding site. Despite the fact that a ligand pentamer was used in crystallization only three of the five sites in the binding site was occupied by a visible glucose unit (and one of these was only partly occupied). Overall, we propose that structural flexibility and a crucial CH–π interaction provided by phenylalanine 110 allows this CBM to retain glucan-based ligands in the bindning site, while leucine in that position establishes a more ligand-restrictive bindning site. (Abstract of the article) |
| 4BJ0 | 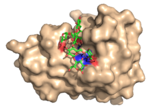 | Structure of the evolved, carbohydrate-crossreactive CBM (X-2 L110F) ligated to a branched oligosaccharide. To accommodate the branches of the ligand several residues of the module are displaced (in relation to the unligated CBM (PDB: 2Y6H)). Furthermore, several polar interactions made with linear ligands are lost while new polar interactions, in particular with branching xyloses are made. Several prominent CH-π interactions seem to play an important role in the interaction. This study illustrates how a branched ligand can be allowed to fit into a bindning site that otherwise accommodates linear oligomers. (Abstract of the article) |
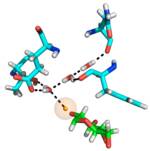
| 5DPN |
Water-mediated hydrogen-bonded network (only some residues are shown). Protein carbons are shown in cyan, ligand carbons in green. Ligand hydrogen/deuterium participating in the network is highlighted in orange. | Structure of the evolved, carbohydrate-crossreactive CBM (X-2 L110F) ligated to a branched oligosaccharide solved by X-ray and neutron diffraction, allowing for excellent views of hydrogen bonds between the protein, the ligand, and water molecules present within the binding site. Several conserved water molecules could be defined as forming a hydrogen-bonded network within the binding site. The protonation stage of a histidine residue that provides critical hydrogen bonding to the ligand was defined and a critical hydrogen bond between the protein and a branch of the ligand was clearly visible. (Abstract of the article). Neutron crystallography offers unique views of protein ligand interactions but certainly poses specific problems in terms of the crystallization process as very large crystals are required for data collection. A process for preparation of such crystals were consequently developed prior to successful data collection. (Abstract of the article) |