Human IgE repertoires and an anti-allergome resource
Background and aim
IgE is the molecular form of antibodies that when bound to mast cells will initiate allergic, hypersensitivity type I, reactions upon encounter with the allergen against which it is specific. Little is known about the molecular nature of antibody recognition of individual allergens. This project aims at defining the characteristics of human IgE recognition of allergens and to provide a resource of human antibodies targeting such allergens for a variety of applications.
Contents
- Approach
- The human IgE-encoding transcriptome
- Allergen-specific repertoires in the IgE population
- Phl p 5 - an important target for allergen-specific IgE
- A common idiotype in IgE repertoires
- Application of human recombinant IgE - hypoallergens
- A human IgE Bet v 1-specific variable domain structure
- General conclusion
- Populärvetenskaplig sammanfattning (in Swedish)
- References
- Funding sources
Approach
To further the understanding of allergic reactions we have sequenced genes encoding human IgE as it is found in allergic individuals (that is in allergy-encoding transcriptomes). We have also (by phage display technology) isolated sequences encoding allergen-specific IgE and characterized such proteins in order to define the diversity of recognition of allergens as it occurs in human allergy.
The human IgE-encoding transcriptome
- We demonstrated that the IgE-encoding transcriptome was highly diverse with respect to V gene usage and showed evidence of somatic hypermutation similar to that found in repertoires encoding for example IgG (Andréasson et al., 2006).
- We showed that despite its diversity the size of the IgE-encoding transcriptomes in these two allergic individuals was highly restricted, that is just a few clonotypes populated the entire population of IgE-poducing B cells. Each set of related clones had, however, been diversified and routes taken in the evolution of these clonotypes could be followed (Figure 1) (Andréasson et al., 2006).
We demonstrated diversity and somatic hypermutation in IgE repertoires in sinus mucosa of patients diagnosed with non-allergic (as assessed by conventional diagnostic tests) sinusitis involving fungal infection and eosinophilic mucus (Levin et al., 2011).
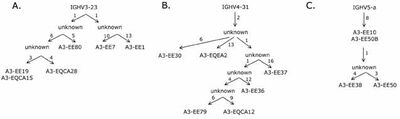
It was shown that very similar rearrangements populated the IgE-encoding transcriptome even in different individuals. Thus, it is likely that for certain epitopes as found on allergens, there are only few ways in which a selectable antigen-binding site can be created based on the available repertoire (Figure 2) (Andréasson et al., 2006). This notion was further explored in a subsequent study (Persson et al., 2008) that demonstrated such similar antibodies targeting the important grass-pollenallergen Phl p 2 (see below).

Allergen-specific repertoires in the IgE population
In order to address the composition of allergen-specific recognition of IgE, we cloned genes encoding heavy chain variable domains as found in the IgE with genes encoding human kappa and lambda light chain sequences. Combinatorial antibody libraries have been made and allergen-specific antibody fragments were selected by phage display technology.
The antibody repertoire targeting the major timothy allergen Phl p 5 has been characterized (Andréasson et al., 2006; Persson et al., 2007).
- Antibody fragments specific for the important thimoty allergen Phl p 5 were selected from our combinatorial libraries. Their sequences demonstarted that they were derived from a diverse set of VH genes. derived from the IGHV1, 3 and 5 subgroups. Thus the response against a single allergen may be complex in geneic terms.
- We demonstrated that specific antibodies against Phl p 5 targeted a range of epitopes on this allergen. At least five such epitopes, three of which are located on a 110 amino acid long fragment of Phl p 5, were identified.
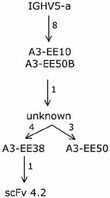
- We were able to put one specific clone, scFv 4.2, into one of the clonotype familes identified above Thus evolutionary paths, as it occurs in vivo, taken by an allergen-specific human clone targeting an biologically relevant, known antigen have been established (Figure 3). Thus by using the toolbox available to us on the IgE-encoding transcriptome, it is possible to define evolutionary paths taken during human immune responses.
To further assess the make-up of the IgE-encoding transcripome, we selected specific binders for additional timothy pollen allergens, namely Phl p 1, Phl p 2, Phl p 4, Phl p 6 and Phl p 11 (Persson et al., 2007).
- We concluded that the response against timothy allergens is diverse and involves genes with an origin in all major VH subgroups (IGHV1, 3, 4 and 5)).
- We defined part of the limited set of antigens that are involved in mounting an IgE response as the 5 investigated timothy allergens accounted for not less than 25% of the Ig-encoding transcriptome of this individual.
- IgE antibodies developed by this phage display approach have recently been shown to be biologically active and to mediate biological activity in the form of antibody-enhanced antigen presentation by dendritic cells that induces a TH2-type immune response (Lundberg et al., 2008).
Altogether, we have developed a unique panel of human IgE specificities targeting six different groups of grass pollen allergens, a probe resource for future assessment of the recognition of allergens by the human humoral immune system. Collectively they represent the first extensive set of an Anti-Allergome Resource, that is sets of IgE representing the human antibody reponse that can interact with the much more extensively characterized Allergome, the set of allergenic proteins that induce type I hypersensitivity reactions by activating effector cells of the immune system that carry IgE.
Phl p 5 - an important target for allergen-specific IgE
Group 5 allergens are major allergens responsible for grass pollen allergy. We have developed and extensively studied (Levin et al., 2014) a large cohort (Andréasson et al., 2006; Levin et al., 2014; Persson et al., 2007) of human IgE targeting an important allergen, Phl p 5 of timothy grass, belonging to this allergen group. We identify at least two largely independent epitopes in both of the domains of this highly flexible allergen. Yet another epitope overlapping two of these epitopes in the N-terminal domain has also been described by us as seen above. There are thus ample opportunities for IgE to cross-link this allergen, a fact that likely contributes to its extensive allergenic potential.
A common idiotype in IgE repertoires
We recently isolated human antibody fragments against the timothy allergen Phl p 2 (Persson et al., 2007) from an antibody library with an origin in the IgE-encoding transcriptome. Interestingly, the sequence of this scFv was very similar to a set of other antibody fragments specific for the same allergen but derived from a library developed from transcripts from another subject (Persson et al., 2008). Furthermore, some transcripts encoding IgE VH in other individuals allergic to grass pollen allergens were very similar in sequence, too. It thus appear that a common sequence motif is involved in the development of specific antibodies targeting a major epitope in the allergen Phl p 2. These findings extend the observation of existence of restricted antibody repertoires (previously defined in e.g. the response against haptens, viral and bacterial antigens, xenoantigens and autoantigens) also to human antibody repertoires targeting allergens.
Application of human recombinant IgE - hypoallergens
With access to human allergens from allergic subjects' repertoires (collections of allergic patients' IgE such as those that we build) we are in a position to utilize the information content of the collections to study allergens and the IgE response in disease and to further develop therapeutic and diagnostic procedures.
A human recombinant IgE holds information that describes, at a clonal level, how such antibodies recognize allergens. W hypothesized that we could use such information to map the epitopes on allergens recognized by human IgE and to modify these epitopes so as to remove IgE recognition, thereby creating a hypoallergenic variant. Such proteins, both naturally occurring and synthetically derived, have been known to exist in the past.
In a recent study (Levin et al., 2013) we demonstrated that it was possible to use a human recombinant IgE collection to develop variants of an allergen, exemplified by the major grass pollen allergen Phl p 1, with IgE-hyporeactive properties. The product can be produced in mg quantities in E. coli, it is folded and it rata ins its ability to induce antibodies able to prevent IgE bindning to wild-type allergen (Levin et al., 2015). We propose that such technology, employing human IgE as a guide in the development of hypoallergens, have the potential to develop new allergen variants with use in specific immunotherapy in allergy.
A human IgE Bet v 1-specific variable domain structure
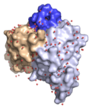
We very recently determined the structure of a human allergen-specific antibody fragment (clone M0418: PDB: 4BUH) in collaboration with researchers at King's College London (Levin et al., 2014). This antibody fragment had been derived from a combinatorial antibody library established from IgE-encoding transcriptome of nasal biopsies. It is very highly specific for an epitope in the tree pollen allergen Bet v 1 and it does not cross-react to a range of other proteins belonging to the PR-10 protein family, a common feature among some antibodies specific for Bet v 1.
This specific binder has a long third complementarity determining region of the heavy chain that forms a loop that protrudes extensively from the surface of the domain creating what appears to be a convex binding site. In this sense it represents a new type of IgE binding site as the only two human IgE variable domain structures that have been determined in the past features flat or concave binding surfaces. This structure suggests that further investigations are needed to define the major binding site features of human IgE, surfaces that may take on more diverse appearances than previously thought.
Epitope deconvolution using peptide microarrays
Immune responses to important allergens may differ for instance in titer and engagement of antibodies of different isotypes. We also hypothesise that it may differ in the complexity of epitopes recognised by the immune system. Definition of epitope recognition at large scale is a difficult challenge. Confrmational epitopes require complex analytical tools (e.g. X-ray crystallography, HDX-MS, Cryo-EM, or extensive mutatgenesis approaches) that have low throughput, We, in collaboration with research teams at KTH, SciLifeLab, and Karolinska Institute, selected to build a comprehensive peptide microarray that covers all (more than 175000) 16-mer linear peptides of all allergens, isoallergens, and isoforms reported in the IUIS allergen database (https://www.allergen.org) (Mikus et al., 2021). This tool was used to assess the complexity of IgE, IgG4, and IgG responses to important allergens of grass and birch pollen (Mikus et al., 2021; Thörnqvist et al., 2022). Large interindividual differences in epitope recognition were observed. Allergen-specific immunotherapy induced recognition of novel epitopes but early responses remained stable during the three year study period.
A subsequent study (Franciskovic et al., 2024) also addressed IgG responses to 19 major fish beta-parvalbulin allergens and variants thereof of 12 fish species, and demonstrated substatial interindividual differences in epitope recognition between individuals. In general, epitope profiles remained stable in a given individual during the three year study period, although examles of emergance of new epitopes occurred.
In all, comprehensive peptide microarrays is a valuable tool to assess extent and dynamics of epitope recognition profiles of large sets of allergens and variants thereof. We envisage that future technology development that allows for analysis also of conformational epitopes at large scale will further grow our understanding of epitope recognition of improtance in allergic disease and contribute to our understanding of how such complexity relate to the disease process and disease symptoms.
General conclusion
We have demonstrated that molecular library technology can be used to unravel molecular details of IgE as it occurs in allergic disease. It can also be used to develop unique research reagents targeting allergens and to develop new approaches to treat and analyze allergic disease. We envisage that such knowledge and reagents will help us understand allergic disease and to improve treatment, diagnosis and prevention of such diseases in the future.
Populärvetenskaplig sammanfattning (på svenska)
Antikroppar är proteiner som oftast skyddar oss mot sjukdom. De gör det genom att binda till främmande ämnen (till exempel bakterier, virus och gifter) och därefter medverka till att dessa förstörs. Tyvärr kan de också i vissa fall ge upphov till sjukdom när fel sorts antikropp reagerar med något ämne på ett olämpligt sätt.
Allergi är en sådan sjukdom, där en speciell sorts antikroppar, så kallat IgE, binder till annars ofta ofarliga ämnen (allergener). Till exempel kan gräspollen, trädpollen, mat (till exempel nötter), djur och kvalster innehålla sådana allergener.
Väldigt många allergener är kända och deras egenskaper är väldigt väl bestämda. Antikroppar, till exempel IgE är däremot mer varierade inte minst därför att varje individ skapar sin egna unika uppsättning av antikroppar. Dessutom finns IgE, i motsats till många andra antikroppar som skyddar oss mot sjukdom, i väldigt små mängder i kroppen. De är därför svåra att komma åt och studera i detalj.
Vi använder oss av molekylärbiologiska metoder för att isolera och studera mänskliga IgE som binder till allergener (till exempel från gräspollen, trädpollen och kvalster). Avsikten är att bättre förstå den allergiska sjukdomen, samt att förbättra diagnostik och behandling av dessa. I det senare fall kan detta ske till exempel genom att möjliggöra framställning av vacciner med mindre grad av biverkning
References
- Andréasson U, Flicker S, Lindstedt M, Valenta R, Greiff L, Korsgren M, Borrebaeck CAK and Ohlin M (2006) The human IgE-encoding transcriptome to assess antibody repertoires and repertoire evolution. J Mol Biol 362, 212-227. (Abstract)
- Persson H, Karbalaei Sadegh M, Greiff L and Ohlin M (2007) Delineating the specificity of an IgE-encoding transcriptome. J Allergy Clin Immunol 120, 1186-1192. (Abstract) (see comment in the journal's Editors' choice section)
- Persson H (2007) Generation and evolution of human antibody repertoires. (doctoral thesis, Lund University) (Abstract)
- Persson H, Flicker S, Karbalaei Sadegh M, Valenta R, Greiff L and Ohlin M (2008) A common idiotype in IgE and its relation to recognition of the grass pollen allergen Phl p 2. Mol Immunol 45, 2715-2720. (Abstract)
- Persson J, Augustsson P, Laurell T and Ohlin M (2008) Acoustic microfluidic chip technology to facilitate automation of phage display selection. FEBS J 275, 5657-5666. (Abstract) (journal cover)
- Lundberg K, Lindstedt M, Larsson K, Dexlin L, Wingren C, Ohlin M, Greiff L and Borrebaeck CAK (2008) Augmented Phl p 5-specific Th2 response after exposure of dendritic cells to allergen in complex with specific IgE compared to IgG1 and IgG4. Clin Immunol 128, 358-365. (Abstract)
- Levin M, Tan LW, Baker L, Wormald PJ, Greiff L, Ohlin M (2011) Diversity of IgE-encoding transcripts in sinus mucosa of subjects diagnosed with non-allergic fungal eosinophilic sinusitis. Clin Exp Allergy 41, 811-820. (Abstract)
- Levin M, Ohlin M (2013) Inconclusive evidence for or against positive antigen selection in the shaping of human IgE repertoires - a call for new approaches. Int Arch Allergy Immunol 161, 122-126. (Abstract)
- Levin M, Rydnert F, Källström E, Tan LW, Wormald PJ, Lindstedt M, Greiff L, Ohlin M (2013) Phl p 1–specific human monoclonal IgE and design of a hypoallergenic group 1 grass pollen allergen fragment. J Immunol 191, 551-560. (Abstract)
- Gadermaier E, Levin M, Flicker S, Ohlin M (2014) The human IgE repertoire. Int Arch Allergy Immunol 163, 77-91. (Abstract)
- Levin M, Davies AM, Liljekvist M, Carlsson F, Gould HJ, Sutton BJ, Ohlin M (2014) Human IgE against the major allergen Bet v 1 – defining an epitope with limited cross-reactivity between different PR-10 family proteins. Clin Exp Allergy 44, 288-299. (Abstract)
- Levin M, Rotthus S, Wendel S, Najafi N, Källström E, Focke-Tejkl M, Valenta R, Flicker S, Ohlin M (2014) Multiple independent IgE epitopes on the highly allergenic grass pollen allergen Phl p 5. Clin Exp Allergy 44, 1409-1419. (Abstract)
- Levin M, Otten H, von Wachenfeldt C, Ohlin M (2015) A folded and immunogenic IgE-hyporeactive variant of the major allergen Phl p 1 produced in Escherichia coli. BMC Biotechnol 15, 52. (Abstract)
- Levin M, King JJ, Glanville J, Jackson KJL, Looney TJ, Hoh RA, Mari A, Andersson M, Greiff L, Fire AZ, Boyd SD, Ohlin M (2016) Persistence and evolution of allergen-specific IgE repertoires during subcutaneous specific immunotherapy. J Allergy Clin Immunol 137, 1535-1544. (Abstract)
- Levin M, Levander F, Palmason R, Greiff L, Ohlin M (2017) Antibody-encoding repertoires produced by cells of bone marrow and peripheral blood - a focus on IgE. J Allergy Clin Immunol 139, 1026-1030. (Abstract)
- Hjort C, Schiøtz PO, Ohlin M, Würtze PA, Christensen LH, Hoffmann HJ (2017) The number and affinity of productive IgE pairs determine allergen activation of mast cells. J Allergy Clin Immunol 140, 1167-1170. (Abstract)
- Glesner J, Kapingidza AB, Godzwon M, Offermann LR, Mueller GA, DeRose EF, Wright P, Richardson CM, Woodfolk JA, Vailes LD, Wunschmann S, London RE, Chapman MD, Ohlin M, Chruszcz M, Pomes A (2019) A human IgE antibody binding site on Der p 2 for the design of recombinant allergen for immunotherapy. J Immunol 203, 2545-2556. (Abstract)
- Mikus M, Zandian A, Sjöberg R, Hamsten C, Forsström B, Andersson M, Greiff L, Uhlén M, Levin M, Nilsson P, van Hage M, Ohlin M (2020) Allergome-wide peptide microarrays enable epitope deconvolution in allergen-specific immunotherapy. J Allergy Clin Immunol 147, 1077-1086. (Abstract at publisher's website)
- Thörnqvist L, Sjöberg R, Greiff L, van Hage M, Ohlin M (2022) Linear epitope binding patterns of grass-pollen specific antibodies in allergy and in response to allergen-specific immunotherapy. Front Allergy 3:859126 (Abstract at publisher's website)
- Hoh RA, Thörnqvist L, Yang F, Godzwon M, King JJ, Lee J-Y, Greiff L, Boyd SD, Ohlin M (2023) Clonal evolution and stereotyped sequences of human IgE lineages in aeroallergenspecific immunotherapy. J Allergy Clin Immunol 152, 214-229. (Abstract at JACI's website)
- Franciskovic E, Thörnqvist L, Greiff L, Gasset M, Ohlin M (2024) Linear epitopes of bony fish β-parvalbumins. Front Immunol (in press) (Abstract at Frontier's website)
This project has been supported by grants from
- Alfred Österlund's Foundation
- the Swedish Research Council
- Stiftelsen Olle Engkvist Byggmästare
- the Konsul Th C Bergh's Foundation
- the Crafoord Foundation
- the Swedish Asthma- and Allergy Association's Research Foundation

