Antigen-designed libraries
A designed antibody library for isolation of hapten-specific binders
Antibody topography
The topography of an antigen-binding site is determined by the length and sequence composition of the CDR loops as well as by the number and positioning of the antigen contact residues. Such topography is strongly correlated to the size of the antigen to which the antibody interacts. Based on these considerations we have designed (Persson et al., 2006) a focused single chain antibody fragment (scFv) repertoire biased for haptens, designated the cavity library. The hapten specific scFv, FITC8, was used as a scaffold for library construction. This scFv, like other hapten binders, display a characteristic cavity in its paratope into which the hapten binds.
Selection strategy
In five of the six CDR-loops, diversity carrying residues were rationally selected based on a model structure of FITC8 and on known antibody structure-function relationships, resulting in variation of 11 centrally located, cavity-lining residues. CDRL3 was allowed to carry a more complex type of diversity. In addition, length variation was introduced in H2 as longer versions of this loop have been shown to correlate with increased hapten binding. Altogether a library with diversity limited to a part of the binding site likely to interact with haptens (Figure 1).
The library was screened, using phage display, against a panel of different haptens, yielding diverse and highly specific binders to four of the antigens. Parallel selections were performed with a library having diversity spread onto a greater area, including more peripherally located residues. This resulted in the isolation of binders, which in contrast to the clones selected from the cavity library, were not able to bind to the soluble hapten in absence of the carrier protein. Thus the designed cavity library behaved better with respect to specificity as compared to a conventional "single-pot" library.
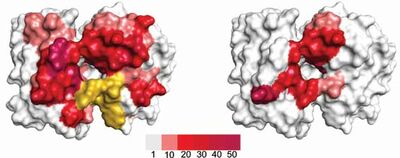
Conclusion
In conclusion, we have shown that by focusing diversity to the hot-spots of interaction a library with improved hapten binding ability can be created. The study also supports the notion that it is possible to create antibody libraries that are biased for the recognition of antigens of pre-defined size. These results also suggest that it is possible to design libraries in a way that allows the derived products to have a better specificity profile as compared to conventional "single-pot" library designs.
Importantly, an analysis has also demonstrated that members selected from such populations are excellent sources to develop high affinity binders following affinity maturation by random mutagenesis and stringent phage display selection (Persson et al. 2008). Indeed, an approach exploiting a selected polyclonal population was used to identify high affinity binders. Mutations in high affinity variants were scattered throughout the scaffold and could not easily have been predicted.
Furthermore, improved variants were isolated that were derived from a range of clones suggesting that a polyclonal origin might be beneficial in the retrieval of high affinity variants as it may be difficult to predict the variants with best potential for affinity evolution in advance. We suggest that polyclonal populations of genes should be used as a source of diversity and into which variability is incorporated to achieve efficient affinity maturation of specific binders.
References
- Persson H, Lantto J and Ohlin M (2006) Creating a focused antibody library for improved hapten recognition. J Mol Biol 357, 607-620. (Abstract at publisher's website)
- Persson H (2007) Generation and evolution of human antibody repertoires. Doctoral thesis. (Abstract at publisher's website)
- Persson H, Wallmark H, Ljungars A, Hallborn J and Ohlin M (2008) In vitro evolution of an antibody fragment population to find high affinity hapten binders. Protein Eng Des Sel 21, 485-493. (Abstract at publisher's website)

