Combinatorial libraries and phage display
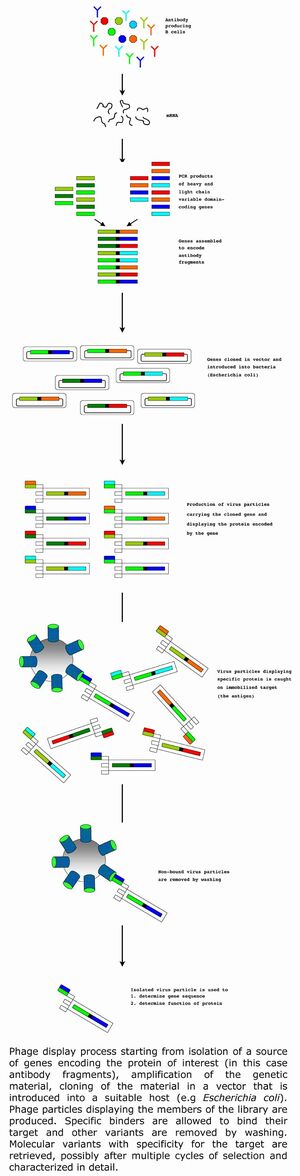
Combinatorial library technology in combination with selection principles like phage display technology has since its introduction in the mid 1980-ies (Smith, 1985) revolutionized the development of biomolecules.
Phage display is an elegant approach whereby the products of a gene harbored within the genome of a bacterial virus (a phage) or a so-called phagemid vector (a plasmid that fools the virus system to believe that it is a viral genome) will be found on the surface of the virus particle. In this way, the genes encoding the protein product will be found inside the virus particle while the function encoded for by the gene (such as the binding properties of an antibody) will be displayed on the surface of the virus.
This combination has two attractive features. Firstly it allows us to select for the properties of the protein on the virus surface. Secondly we can analyze the sequence of the gene found inside the particle that on its surface carried the protein with a desirable property, like binding to a toxin, a cancer cell or a bacterium.
This approach has two important advantages. Firstly, the function (for example the binding properties) of the protein encoded for by the gene cannot be itelf be easily predicted from the gene sequence. Secondly, the sequence of a protein with a desirable function cannot easily be predicted from the function itself. Rather the combined set provides critical complementary sets of information, namely function of the protein and sequence of the gene.
Through the realization that these features could be put together in a single concept as evidenced by phage display (Smith, 1985) we gained access to a tool that allows us to develop proteins with specific binding properties at a rate exceeding that of other technologies.
Initial efforts were focused on the development of peptides with specific binding properties (Smith, 1985). Developments in the field of protein engineering, in particular
- the realization that heterologous complex proteins like antibody fragments can be produced in functional form in E. coli and secreted from the cell using e.g. pelB leader sequences
- the realization that diverse antibody-encoding gene sequences could be amplified by PCR (polymerase chain reaction) using a restricted set of primers (Larrick et al., 1989; Orlandi et al., 1989)
paved the way for similar efforts to identify specific binding proteins from large combinatorial libraries.
Altogether these developments were critical for the work that eventually resulted in the first description of a selection process of displayed proteins on the surface of filamentous bacteriophage (McCafferty et al., 1990).
Since then phage display and other technologies like ribosome display (Hanes and Plückthun, 1997), bacterial display (Georgiou et al., 1997), yeast display (Boder and Wittrup, 1997) have been fine-tuned to deliver proteins with very defined properties. Numerous combinatorial libraries have been made either directed towards defined antigens or larger, "naïve" libraries suitable for essentially any type of target (as exemplified by de Haard et al., 1999; Griffiths et al., 1994; Knappik et al., 2000; Løset et al., 2005; Marks et al., 1994; Nissim et al., 1994; Pini et al., 1998; Sheets et al., 1998; Söderlind et al., 2000; Vaughan et al., 1996).
Proteins selected from antibody libraries displayed on phage particles are already in clinical use (Humira®) and many more are undergoing clinical trials for use in the treatment of e.g. cancer, infectious disease and autoimmune disease. Thus this technology has proven its capacity to deliver proteins of biomedical importance.
References:
- Boder ET and Wittrup KD (1997) Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol 15, 553-557.
- de Haard HJ, van Neer N, Reurs A, Hufton SE, Roovers RC, Henderikx P, de Bruïne AP, Arends JW and Hoogenboom HR (1999) A large non-immunized human Fab fragment phage library that permits rapid isolation and kinetic analysis of high affinity antibodies.
J Biol Chem 274, 18218-18230. - Georgiou G, Stathopoulos C, Daugherty PS, Nayak AR, Iverson BL and Curtiss R III (1997) Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines. Nat Biotechnol 15, 29-34.
- Griffiths AD, Williams SC, Hartley O, Tomlinson IM, Waterhouse P, Crosby WL, Kontermann RE, Jones PT, Low NM, Allison TJ, et al. (1994) Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J 13, 3245-3260.
- Hanes J and Plückthun A (1997) In vitro selection and evolution of functional proteins by using ribosome display. Proc Natl Acad Sci USA 94, 4937-4942.
- Knappik A, Ge L, Honegger A, Pack P, Fischer M, Wellnhofer G, Hoess A, Wölle J, Plückthun A and Virnekäs B (2000) Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J Mol Biol 296, 57-86.
- Larrick JW, Danielsson L, Brenner CA, Abrahamson M, Fry KE and Borrebaeck CAK (1989) Rapid cloning of rearranged immunoglobulin genes from human hybridoma cells using mixed primers and the polymerase chain reaction. Biochem Biophys Res Commun 160, 1250-1256.
- Løset GA, Løbersli I, Kavlie A, Stacy JE, Borgen T, Kausmally L, Hvattum E, Simonsen B, Hovda MB and Brekke OH (2005) Construction, evaluation and refinement of a large human antibody phage library based on the IgD and IgM variable gene repertoire. J Immunol Methods 299, 47-62.
- Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD and Winter G (1994) By-passing immunization. Human antibodies from V-gene libraries displayed on phage.
J Mol Biol 222, 581-597. - McCafferty J, Griffiths AD, Winter G and Chiswell DJ (1990) Phage antibodies: filamentous phage displaying antibody variable domains. Nature 348, 552-554.
- Nissim A, Hoogenboom HR, Tomlinson IM, Flynn G, Midgley C, Lane D and Winter G (1994) Antibody fragments from a 'single pot' phage display library as immunochemical reagents. EMBO J 13, 692-698.
- Orlandi R, Gussow DH, Jones PT and Winter G (1989) Cloning immunoglobulin variable domains for expression by the polymerase chain reaction. Proc Natl Acad Sci USA 86, 3833-3837.
- Pini A, Viti F, Santucci A, Carnemolla B, Zardi L, Neri P and Neri D (1998) Design and use of a phage display library. Human antibodies with subnanomolar affinity against a marker of angiogenesis eluted from a two-dimensional gel. J Biol Chem 273, 21769-21776.
- Sheets MD, Amersdorfer P, Finnern R, Sargent P, Lindquist E, Schier R, Hemingsen G, Wong C, Gerhart JC and Marks JD (1998) Efficient construction of a large nonimmune phage antibody library: the production of high-affinity human single-chain antibodies to protein antigens. Proc Natl Acad Sci USA 95, 6157-6162.
- Skerra A and Plückthun A (1988) Assembly of a functional immunoglobulin Fv fragment in Escherichia coli. Science 240, 1038-1041.
- Smith GP (1985) Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228, 1315-1317.
- Söderlind E, Strandberg L, Jirholt P, Kobayashi N, Alexeiva V, Åberg AM, Nilsson A, Jansson B, Ohlin M, Wingren C, Danielsson L, Carlsson R and Borrebaeck CAK (2000) Recombining germline-derived CDR sequences for creating diverse single-framework antibody libraries. Nat Biotechnol 18, 852-856.
- Vaughan TJ, Williams AJ, Pritchard K, Osbourn JK, Pope AR, Earnshaw JC, McCafferty J, Hodits RA, Wilton J and Johnson KS (1996) Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat Biotechnol 14, 309-314.
