Identification of biomarkers for ovarian cancer
Biomarkers for ovarian cancer
Ovarian cancer affects roughly 1,5 % of the female population and is the most fatal of gynaecological cancers. Currently, the disease is diagnosed through a combination of pelvic exams, ultrasound or CT scans and low-specificity blood tests but frequently a firm diagnosis can only be set after invasive surgery. Treatment options include surgery to remove one, or both ovaries, sometimes in combination with the uterus as well as chemotherapy and targeted immune-oncology approaches. Interventions involving surgery are associated with patient discomfort and, in some cases, reproductive failure.
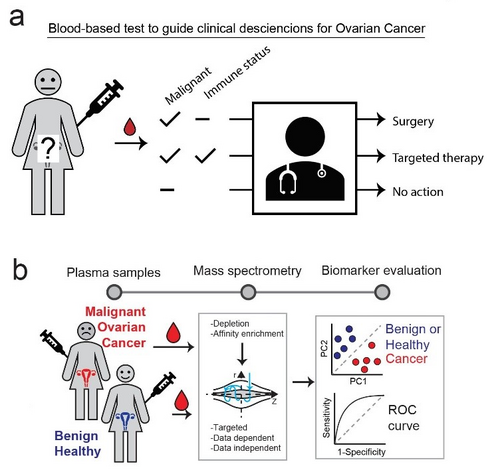
We aim to develop a non-invasive blood-based test that discriminates benign and healthy from malignant conditions in addition to guiding clinical decisions regarding choice of targeted immune-modulatory therapy (Fig 1a). To this end we perform comprehensive analysis of plasma proteome from malignant and control conditions using quantitative mass spectrometry-based proteomics (Fig 1b). This allows us to globally explore correlations between plasma protein levels and clinical parameters such as disease state, histological grade etc.
Aside from developing precision diagnostic tools for ovarian cancer, we also explore fundamental molecular mechanisms to identify potential new drug targets.
A recent highlight includes the characterization of the human enzyme METTL13 which was initially reported to be over-represented in the plasma of patients with high grade serous ovarian cancers (HGSC) [1]. We uncovered METTL13 as a methyltransferase regulating protein synthesis through methylation of a translational elongation factor [2]. METTL13-mediated increased translational output was subsequently reported promote tumorigenesis [3]. Collectively these findings were acknowledged as a Research Highlight in Nature Chemical Biology [4].
Previous research efforts have focused on a specific lysine methylation event on the human stress-inducible 70 kDa heat shock protein (Hsp70-K561). Through molecular biology studies using gene targeted cells and proteomics we uncovered human METTL21A to be exclusively responsible for this PTM [5,6] and demonstrated that the degree of methylation at Hsp70-K561 in HGSC effusions represents a prognostic biomarker associated with patient survival [7].
References
[1] Li, Y. et al. Immunogenic FEAT protein circulates in the bloodstream of cancer patients. J. Transl. Med. 14, 1–12 (2016).
[2] Jakobsson, M. E. et al. The dual methyltransferase METTL13 targets N terminus and Lys55 of eEF1A and modulates codon-specific translation rates. Nat. Commun. 9, 3411 (2018).
[3] Liu, S. et al. METTL13 Methylation of eEF1A Increases Translational Output to Promote Tumorigenesis. Cell 176, 491-504.e21 (2019).
[4] Miura, G. METTLing with translation. Nat. Chem. Biol. 15, 207 (2019).
[5] Jakobsson, M. E., Moen, A., Davidson, B. & Falnes, P. Hsp70 (HSPA1) lysine methylation status as a potential prognostic factor in metastatic high-grade serous carcinoma. PLoS One 10, 1–13 (2015).
[6] Jakobsson, M. E., Moen, A. & Falnes, P. O. Correspondence: On the enzymology and significance of HSPA1 lysine methylation. Nat Commun 7, 11464 (2016).
[7] Jakobsson, M. E. et al. Identification and characterization of a novel human methyltransferase modulating Hsp70 protein function through lysine methylation. J. Biol. Chem. 288, 27752–27763 (2013).
