Development of methodology for PTM proteomics
PTM proteomics
The function of proteins is frequently regulated by dynamic post translational modifications (PTMs). Prominent examples include specific glycosylation events that define blood-type, phosphorylation events important in cell signalling and methylation which is a key epigenetic mark playing a central role in gene regulation. We have a long-standing interest in the characterization of PTMs and develop proteomics technology for this purpose.
Lysine methylation
A key research focus of the lab is the enzymology and function of lysine methylation (Kme). Kme is a dynamic PTM and is regulated by both methyltransferase (writing) and demethylase (erasing) enzymes (Fig 1a). Moreover, Kme interacting domains (readers) exist that can mediate functions governed by the PTM (Fig 1b).
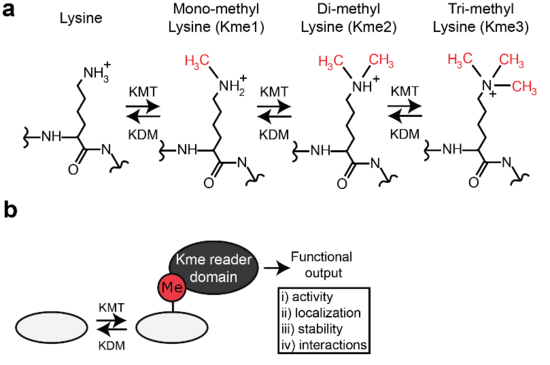
Figure 1. Biochemistry and function of lysine methylation.
(a) Chemical structure of the different methylated forms of lysine.
(b) Diverse functions of lysine methylation can be mediate by specific reader domain containing proteins.
The method of choice for large scale analysis of PTMs is mass spectrometry (MS). PTMs are typically of low abundance and are enriched by specific affinity agents prior to MS analysis. For Kme, there are no affinity agents readily available and consequently the function and biology of Kme are scientifically underexplored.
We aim to develop new and robust affinity agents for MS-based proteomic characterization of Kme (Fig 2a). To this end we currently employ three strategies comprising i) cloning and isolation of recombinant Kme reader domains encoded in mammalian genomes, ii) selection of Kme readers through phage display of recombinant antibody libraries and iii) traditional murine immunizations.
The functional Kme binders are subsequently used to explore the context and abundance of the PTM in cells and tissues (Fig 2b-d) as well as in biomarker discovery studies (Fig 2e-f).
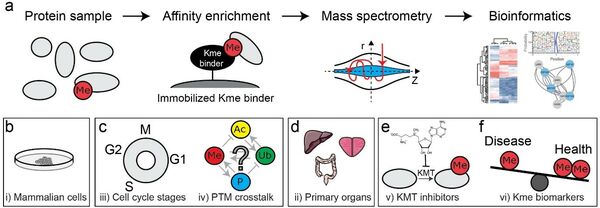
Figure 2. Affinity proteomics mediated exploration of lysine methylation.
(a) Workflow for Kme proteomics analysis.
(b-f) applications of Kme proteomics.
References
Jakobsson ME* et al. The dual methyltransferase METTL13 targets N terminus and Lys55 of eEF1A and modulates codon-specific translation rates. Nat. Commun. 9, 3411 (2018).
*shared corresponding author
Jakobsson ME et al.. Regulation of eukaryotic elongation factor 1 alpha (eEF1A) by dynamic lysine methylation. RNA Biol. 6286, 01–11 (2018).
Willemen HLDM, Kavelaars A, Prado J, Maas M, Versteeg S, Nellissen LJJ, Tromp J, Gonzalez Cano R, Zhou W, Jakobsson ME et al.. Identification of FAM173B as a protein methyltransferase promoting chronic pain. PLOS Biol. 16, e2003452 (2018).
Małecki J, Jakobsson ME et al. Uncovering human METTL12 as a mitochondrial methyltransferase that modulates citrate synthase activity through metabolite-sensitive lysine methylation. J. Biol. Chem. 292, 17950–17962 (2017).
Jakobsson ME* et al. Methylation of human eukaryotic elongation factor alpha (eEF1A) by a member of a novel protein lysine methyltransferase family modulates mRNA translation. Nucleic Acids Res. 45, 8239–8254 (2017).
*shared corresponding author
Malecki J, Aileni VK, Ho AYY, Schwarz J, Moen A, Sørensen V, Nilges BS, Jakobsson ME et al. The novel lysine specific methyltransferase METTL21B affects mRNA translation through inducible and dynamic methylation of Lys-165 in human eukaryotic elongation factor 1 alpha (eEF1A). Nucleic Acids Res. 45, 4370–4389 (2017).
Falnes PØ, JakobssonME et al. Protein lysine methylation by seven-β-strand methyltransferases. Biochem. J. 473, 1995–2009 (2016).
Jakobsson MEet al. Correspondence: On the enzymology and significance of HSPA1 lysine methylation. Nat Commun 7, 11464 (2016).
Jakobsson ME. et al. Hsp70 (HSPA1) lysine methylation status as a potential prognostic factor in metastatic high-grade serous carcinoma. PLoS One 10, 1–13 (2015).
Jakobsson ME* et al. Saccharomyces cerevisiae eukaryotic elongation factor 1A (eEF1A) is methylated at Lys-390 by a METTL21-like methyltransferase. PLoS One 10, 1–18 (2015).
*shared corresponding author
Davydova E, Ho AY, Malecki J, Moen A, Enserink JM, Jakobsson ME et al. Identification and characterization of a novel evolutionarily conserved lysine-specific methyltransferase targeting eukaryotic translation elongation factor 2 (eEF2). J. Biol. Chem. 289, 30499–30510 (2014).
Jakobsson ME et al. Identification and characterization of a novel human methyltransferase modulating Hsp70 protein function through lysine methylation . J. Biol. Chem.288, 27752-63 (2013).
Kernstock, S., Davydova, E., Jakobsson ME et al. Lysine methylation of VCP by a member of a novel human protein methyltransferase family. Nat. Commun. 3, 1038 (2012)
